Mouse strains
All procedures were performed with 8–10-week-old C57BL/6 Slc11a1+/+, Slc11a1D169/D169, Slc11a1+/+ Cepbe+/+, Slc11a1+/+ Cepbe−/−, Slc11aD169/D169 Cepbe+/+ or Slc11aD169/D169 Cepbe−/− mice. Male and female mice were used for each experiment unless otherwise specified in the figure legend. Slc11a1+/+ mice were obtained from G. Barton22, rederived into the barrier facility at UC Davis, backcrossed once with C57BL/6J mice and then bred and maintained under specific pathogen-free conditions by the UC Davis Teaching and Research Animal Care Service. Specific pathogen-free Slc11a1D169/D169 mice were purchased from The Jackson Laboratory (Bar Harbor, Maine). In addition, timed-pregnant female Slc11a1D169/D169 mice were purchased from The Jackson Laboratory and the pups were raised in the barrier facility at UC Davis to obtain VAD Slc11a1D169/D169 mice. Cebpe mice37 were provided by H. Phillip Koeffler and were backcrossed onto C57BL/6J and C57BL/6J-Slc11a1+/+ mice. All mice were held in microisolator cages with sterile ALPHA-dri bedding and received irradiated rodent feed and sterile drinking water ad libitum. The UC Davis Institutional Animal Care and Use Committee approved all animal experiments described in this paper under protocols 22492 and 23360.
VAD mice and mice with sufficient vitamin A
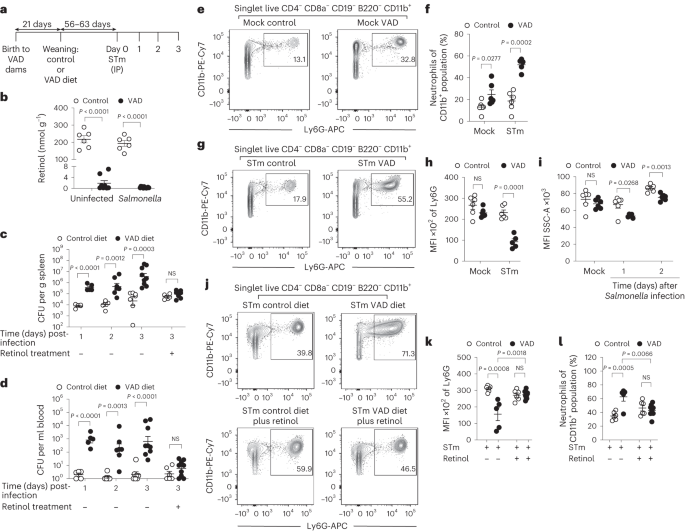
Starting at 14 days of gestation, dams were fed a VAD diet (0 IU vitamin A kg−1). At weaning, mice were either placed on a diet replete with vitamin A (4,000 IU vitamin A kg−1) or maintained on the VAD diet. All diets were semi-purified and casein based. A custom VAD diet (TD.88407) and a vitamin A control diet with added orange food colouring (TD.09062) were prepared and pelleted by Envigo Teklad Diets. For vitamin A supplementation, VAD and control mice were treated with either 0.1 ml of sterile phosphate-buffered saline (PBS) or 600 IU retinyl palmitate (Nutrisorb A, Interplexus) in 0.1 ml of PBS by oral gavage at 7 and 3 days before infection with S. Typhimurium. For vitamin A treatment, VAD and control mice were treated with either 0.1 ml of sterile PBS or 600 IU retinyl palmitate in 0.1 ml of PBS by oral gavage starting 1 day after S. Typhimurium infection.
Measurement of hepatic vitamin A levels
About 20 mg (control) to 50 mg (VAD) of the liver were homogenized with 100 mg sodium sulfate. The homogenate was transferred into a 7 ml glass vial and mixed with 350 µl ethanol, containing 0.1% butylated hydroxytoluol, for protein precipitation. After 100 µl of potassium hydroxide (KOH, 30% in deionized water) and 50 µl pyrogallol (10% in ethanol) were added, the samples were mixed for 15 s and incubated for 60 min at 60 °C to release the retinol from its retinyl esters. After the samples were cooled on ice, 3 ml hexane, 1 ml tocol (internal standard; 1 µg ml−1 in hexane) and 700 µl deionized water were added, mixed vigorously for 30 s and centrifuged for 2 min at 1,800 rpm for phase separation. The upper, organic phase was transferred into a fresh 7 ml glass vial and evaporated to dryness under a gentle nitrogen stream. The residue was reconstituted in 100 µl (VAD liver) or 200 µl (control liver) of acetonitrile before analysis. The analysis was carried out using an Agilent 1100 HPLC system equipped with a diode array detector (Agilent) controlled by OpenLABS ChemStation software (Rev A.01.04, Agilent). Samples were kept at 10 °C, and 10 µl was injected onto a Spherisorb ODS2 column, 125 × 3 mm, 3 µ (Waters) protected by a BDS-hypersil-C18 guard column, 20 × 3 mm, 3 µ (Thermo Scientific) at 15 °C. Acetonitrile, dichloromethane and methanol (7/2/1, v/v/v, all HPLC grade) served as isocratic mobile phase at a flow rate of 0.6 ml min−1 for 6 min. The detector was set at 325 nm for retinol and 295 nm for tocol, and quantification was carried out by ratio response to the internal standard.
LPS challenge
Lipopolysaccharide from E. coli 0111:B4 strain (Invivogen catalogue code tlrl-eblps) was diluted in sterile, 0.9% sodium chloride, and 20 µg per mouse was administered intraperitoneally in a final volume of 100 µl. Male and female mice on control (n = 5) or VAD (n = 6) diets were killed 2 h after injection. Blood samples were collected in K2 ethylenediaminetetraacetic acid (EDTA) tubes, and plasma was collected and stored at −80 °C until further analysis. Cytokine levels were assessed with enzyme-linked immunosorbent assays for TNF-α (BioLegend catalogue number 430904) and IL-6 (Invitrogen catalogue number 88-7064).
S. Typhimurium
A derivative of the S. Typhimurium clinical isolate D23580 NalR (gyrA S83F) pSLT-14028s::tetRA, designated JK1128, was provided by F. Fang and was used for animal infection studies20. Mice received either 0.1 ml of sterile PBS or 1 × 103 colony-forming units (CFU) diluted in PBS by IP injection. For oral infection, mice received either 0.1 ml of Luria–Bertani (LB) broth or 1 × 109 CFU diluted in LB broth by IG gavage. Inocula were cultured for 16–18 h aerobically at 37 °C. To determine tissue loads of viable S. Typhimurium, liver and spleen tissues were homogenized in PBS using an Ultra Turrax T25 basic mixer (IKA). Blood was collected by cardiac puncture with heparinized needles; plasma was removed and then incubated with 120 μl of 1% Triton X-100 in PBS for 10 min at room temperature. Homogenates were serially diluted and plated on LB agar plates containing 100 mg l−1 nalidixic acid (Sigma). After overnight growth at 37 °C, CFU g−1 or CFU ml−1 was calculated.
RNA extraction, reverse-transcription PCR and real-time PCR
For whole-tissue RNA extractions, samples were snap-frozen in liquid nitrogen at time of necropsy and stored at −80 °C. RNA isolation from purified bone marrow neutrophils was performed on the same day. RNA was extracted from samples using Tri-Reagent (Molecular Research Center) according to the manufacturer’s instructions. All RNA samples were treated with DNase I (Ambion) to remove genomic DNA contamination. For quantification of messenger RNA (mRNA) levels in spleen tissue, 1 μg of total RNA from each sample was reverse transcribed in a 50 μl volume (TaqMan reverse transcription (RT) reagent; Applied Biosystems), and 4 μl of the resulting complementary DNA (cDNA) was used for each real-time reaction. For mRNA quantification from purified bone marrow neutrophils, 0.8 μg of total RNA from each sample was reverse transcribed in a 50 μl volume (TaqMan RT reagent; Applied Biosystems) and 4 μl of the resulting cDNA was used for each real-time reaction. Real-time PCR was performed using the primers listed in Table 1, SYBR green (Applied Biosystems) and ViiA 7 Real-Time PCR System (Applied Biosystems). Target gene transcription of each sample was normalized to the respective levels of β-actin (ACTB) or 18S rRNA, and absolute quantification was determined using gene-specific plasmid standards in each run.
Western blot
Protein was extracted from bone marrow neutrophils of control and VAD mice using Tri-Reagent (Molecular Research Center) according to the manufacturer’s instructions. The concentration of bone marrow neutrophil protein was measured using a modified Bradford assay. Briefly, samples were diluted in 0.15 M NaCl and 1 ml of Bradford substrate (0.1 mg ml−1 Coomassie Brilliant Blue G-250, 5% ethanol and 10% of 85% (w/v) phosphoric acid) was added to 100 μl of either sample of standard. A 10 μg sample of protein was separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane (Millipore). A blocking solution of 2.5% non-fat dried milk and 0.1% Tween 20 (Bio-Rad) in PBS was used. For lactoferrin detection, a 1:200 dilution of the primary antibody (lactoferrin (H-65) rabbit polyclonal IgG, catalogue number sc-25622, Santa Cruz Biotechnology) in blocking solution was added to the membrane. As a loading control, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was detected at a 1:5,000 dilution of the primary antibody (GAPDH rabbit mAb, clone 14C10, catalogue number 2118S, Cell Signaling) in blocking solution. A goat anti-rabbit horseradish peroxidase conjugated secondary antibody (Bio-Rad) was diluted 1:3,000 in blocking buffer and applied to the membrane. All antibodies were validated by the manufacturers. Protein bands were visualized by chemiluminescence (SuperSignal West Femto Maximum Sensitivity Substrate, ThermoFisher Scientific) using a BioSpectrum (UVP) imaging system. Raw images were processed using Photoshop CS2 (Adobe Systems) to uniformly adjust brightness.
Isolation of neutrophils from mouse bone marrow
Bone marrow fractionation was performed using a modification of the density gradient centrifugation method previously described49. Briefly, bone marrow was flushed from the femora and tibiae with 10 ml of sterile PBS and passed through an 18-gauge needle to disrupt larger bone marrow clumps. Cells were centrifuged at 300 × g for 7 min at 4 °C. Red blood cells were lysed by resuspending a cell pellet in 0.2% NaCl for 20 s followed by the addition of 1.6% NaCl. Cells were centrifuged at 300 × g for 7 min at 4 °C, washed with 2 mM EDTA in PBS and filtered through a 40 μm filter. Using a 15 ml conical tube, 3 ml of Histopaque 1119 (density 1.119 g ml−1, Sigma-Aldrich) was overlaid with 3 ml of Histopaque 1077 (density 1.077 g ml−1, Sigma-Aldrich). Bone marrow cells were resuspended in 1 ml of ice-cold PBS and laid over the Histopaque gradient. Samples were centrifuged for 30 min at 700 × g at 25 °C without a break. Neutrophils were collected at the interface of the Histopaque 1119 and Histopaque 1077 layers and then washed twice with PBS and used for further experiments. The composition of the cell population was confirmed using microscopy to have neutrophil morphology as determined by Giemsa staining.
Spleen neutrophil enrichment
Neutrophils were enriched from the spleen of mice infected with S. Typhimurium using the EasySep Mouse Neutrophil Enrichment Kit (STEMCELL Technologies) according to the manufacturer’s protocol. Briefly, spleens were removed aseptically and smashed using a syringe plunger to produce a single-cell suspension. Red blood cells were lysed by the addition of ACK lysing buffer (Lonza). Cells were centrifuged at 600 × g for 10 min at 4 °C, washed twice with Dulbecco’s PBS (dPBS) and filtered using a 70 mm filter. Neutrophils were enriched by immunomagnetic negative selection. For S. Typhimurium counts, purified neutrophils were enumerated from each sample and neutrophils were lysed with 1% Triton X-100 in PBS or radioimmunoprecipitation assay buffer for 10 min at room temperature. The suspension was serially diluted and plated on LB agar plates containing 100 mg l−1 nalidixic acid (Sigma-Aldrich). After overnight growth at 37 °C, bacterial counts were calculated as CFU per 105 or CFU per 5 × 105 neutrophils.
Flow cytometry
Flow cytometry analysis was performed for the detection of neutrophils in the spleen and bone marrow of control and VAD mice mock infected and infected with S. Typhimurium. Single-cell suspensions of spleen and bone marrow tissue were obtained as described previously. Cells were resuspended in 2 ml of dPBS and stained with an Aqua Live/Dead cell discriminator (Invitrogen) according to the manufacturer’s protocol. After Live/Dead staining, cells were washed with dPBS and resuspended in 50 μl of PBS containing 1% bovine serum albumin and 2 mM EDTA (fluorescence-activated cell sorter (FACS) buffer). Cells were stained with an Fc receptor blocking antibody, anti-CD16/32 (93) (eBioscience), for 5 min at 4°C and then stained for 20 min at 4°C with a cocktail of anti-B220 (RA3-6B2) PerCp-Cy5.5, anti-CD19 (6D5) PerCp-Cy5.5, anti-CD8a (53–6.7) PerCyp-Cy5.5, anti-CD4 (RM4-5) PerCp-Cy5.5, anti-CD11b (M1/70) PE-Cy7, anti-Ly6G (1A8) APC and anti-Ly6C (HK1.4) Pacific Blue (all BioLegend). In addition, for bone marrow samples, an anti-Ter119 (TER-119) PerCp-Cy5.5 (BioLegend) was added. All antibodies were validated by the manufacturers. Cells were washed twice in FACS buffer, fixed with BD Cytofix (BD Biosciences) for 30 min at 4 °C and resuspended in FACS buffer. For quantification of cell populations, 50 μl of SPHERO AccuCount Fluorescent Particles 10.1 μm (Spherotech) was added to each sample before analysis. Calculation of absolute counts was performed according to the manufacturer’s protocol. Flow cytometry analysis was performed using a BD (Becton Dickinson) LSRII, and 1.0 × 106 events were collected per mouse. Data were analysed using FlowJo software (BD Biosciences), and gates were based on fluorescence-minus-one controls.
ELISA
The levels of IL-6 and TNF-α in serum samples from control and VAD mice were determined by enzyme-linked immunosorbent assay (ELISA) (R&D Systems), according to the manufacturer’s instructions. The ELISA test was read at 450 nm with an ELISA microplate reader (Bio-Rad Model 680). Data points are the averages of duplicate dilutions.
In vivo depletion of neutrophils
For neutrophil depletion experiments, control and VAD male mice were injected intraperitoneally 1 day before and after S. Typhimurium infection with either 500 μg of rat anti-mouse Ly6G monoclonal antibody, clone 1A8 (BioXCell), or 500 μg of rat IgG2a isotype control, clone 2A3 (BioXCell), diluted in 0.2 ml of dPBS. Neutrophil depletion in the tissue was confirmed by flow cytometry as described previously.
Adoptive transfer
Bone marrow neutrophils were isolated from male and female 8–12-week-old, healthy C57BL/6 Slc11a1+/+ mice using the protocol described above. Once collected, neutrophils were washed twice with dPBS, counted and suspended in dPBS at a concentration of 1.5–2 × 107 cells ml−1. Male VAD mice were inoculated intragastrically 1 day after S. Typhimurium infection with either 0.2 ml of the neutrophil suspension (total of 3–4 × 106 neutrophils per mouse) or 0.2 ml of dPBS. Animals were necropsied 3 days after S. Typhimurium infection to assess bacteriology.
Statistical analysis
The statistical significance of differences between groups was determined by a one- or two-tailed Student’s t-test, or one-way analysis of variance (ANOVA) with a post hoc Tukey or Sidak’s test on logarithmically or arc-sin-transformed data. A P value of 0.05 or less was considered to be significant. Animals were excluded if they were not confirmed to be infected after intraperitoneal (IP) administration of S. Typhimurium. Data points that were identified as outliers were excluded based on the ROUT method. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications23. GraphPad Prism 6 was used to perform analyses (GraphPad).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.