Mice
All mouse experiments were approved by the MSKCC and/or CSHL Internal Animal Care and Use Committee (animal protocol 11-06-011 at MSKCC and 21-4 at CSHL). All relevant animal use guidelines and ethical regulations were followed. Mice were maintained under specific pathogen-free conditions. Housing was on a 12-h–12-h light–dark cycle under standard temperature and humidity of approximately 18–24 °C and 40–60%, respectively. The following mice were used: 3- to 4-month-old C57BL/6 mice (purchased from Charles River), 18-month-old C57BL/6 mice (obtained from the National Institute of Aging) and 6-week-old B6.SJL-Ptrca/BoyAiTac (CD45.1) mice (purchased from Taconic). Mice of both sexes were used at 8–12 weeks of age and 18–20 months of age for the aging experiments, males of 8–12 weeks old for the HFD experiments and females of 6–10 weeks old for T cell isolation. Mice were kept in group housing. Mice had free access to food and water except during the starvation period before glucose or insulin tolerance testing. Aging mice were fed a normal diet (PicoLab Rodent Diet 20, LabDiet), mice on the HFD experiments were fed an HFD (TD.06414, 60% of kcal from fat; Envigo).
Flow cytometry
For in vivo sample preparation, livers were dissociated using the MACS liver dissociation kit (Miltenyi Biotec, 130-1-5-807), filtered through a 100-μm strainer and washed with PBS, and red blood cells were lysed by an ammonium–chloride–potassium (ACK) lysing buffer (Lonza). Cells were washed with PBS, resuspended in FACS buffer and either used for immediate analysis or fixed with Fixation Buffer (BD Biosciences, 554655) according to the manufacturer’s instructions and used for later analysis. Spleens were mechanically disrupted with the back of a 5-ml syringe, filtered through a 40-μm strainer and washed with PBS and 2 mM EDTA; then red blood cells were lysed by ACK lysing buffer (Lonza). Gonadal adipose tissue was dissociated as described42. In short, adipose tissue was isolated and placed in a digestion solution consisting of 4 mg ml−1 collagenase, type II (Sigma) in DPBS (Life Technologies) supplemented with 0.5% BSA (Sigma) and 10 mM CaCl2 digested at 37 °C for 20 min in a rotational shaker. Afterwards, samples were mechanically dissociated with a 10-ml serological pipette, filtered through a 40-μm strainer and washed with PBS and 2 mM EDTA; then red blood cells were lysed by ACK lysing buffer (Lonza). Pancreata were placed into cold DMEM with 10% FBS and penicillin and streptomycin. The pancreata were minced in this media on ice into 2- to 4-mm fragments so that they would pass through the end of a 1-ml pipette tip with ease. The minced tissue was collected in a 15-ml Falcon tube and dissociated in 100 mg ml−1 Dispase (Life Technologies, 17105041), 20 mg ml−1 collagenase P (Roche, 11249002001) and 1 mM EDTA for 20 min on a heated rocker at 37 °C (Eppendorf). After 20 min, 5 ml of DMEM with 10% FBS was added to quench the reaction. The supernatant was removed and filtered through a 100-µm filter (VWR). Next, 5 ml of dissociation media consisting of 100 mg ml−1 Dispase (Life Technologies, 17105041), 20 mg ml−1 collagenase P (Roche, 11249002001) and 1 mM EDTA was added before replacing the 15-ml tube into the heated rocker for 20 min. The reaction was quenched again after 20 min with media and filtered via a 100-µm filter. The dissociated cells were spun at 500 r.c.f. for 10 min in a swinging-bucket centrifuge. The supernatant was discarded, and the cells were resuspended in ACK lysis buffer for 2–4 min in ice. Cells were washed with PBS, resuspended in FACS buffer and either used for immediate analysis or fixed with Fixation Buffer (BD Biosciences, 554655) and used for later analysis.
Fc receptors were blocked using FcR blocking reagent, mouse (Miltenyi Biotec). The following fluorophore-conjugated antibodies were used in the indicated dilutions: Myc-tag AF647 (clone 9B11, Cell Signaling Technology, 2233S, 25; 1:50 dilution), m.CD45.1 BV785 (clone A20, BioLegend, 110743, B347719; 1:100 dilution), m.CD45.2 BV785 (clone 104, BioLegend, 109839, B343292; 1:100 dilution), mCD3 AF488 (clone 17A2, BioLegend, 100210, B284975; 1:100 dilution), mCD4 BUV395 (clone GK1.5, BD, 563790, 1097734; 1:50 dilution), mCD8 PE-Cy7 (clone 53-6.7, BioLegend, 100722, B312604; 1:50 dilution), mCD62L BV421 (clone MEL-14, BioLegend, 104435, B283191; 1:50 dilution), mCD44 APC-Cy7 (clone IM7, BD Pharminogen, 560568, 1083068; 1:100 dilution), mCD3 BV650 (clone 17A2, BioLegend, 100229, B350667; 1:100 dilution), mCD19 BV650 (clone 1D3, BD Biosciences, 563235, 1354015; 1:100 dilution), mNKp46 BV650 (clone 29A1.4, BioLegend, 137635, B298809; 1:100 dilution), mCD11b BUV395 (clone M1/70, BD Biosciences, 563553, 0030272; 1:50 dilution), mLy-6C APC-Cy7 (clone HK1.4, BioLegend, 128026, B375238; 1:100 dilution), mly6G BV605 (clone 1A8, BD Biosciences, 563005, 2144780; 1:100 dilution), m.uPAR AF700 (R&D systems, FAB531N, 1656339; 1:50 dilution), m.uPAR PE (R&D systems, FAB531P, ABLH0722051; 1:50 dilution), mF4/80 PE-eFluor610 (clone BM8, Invitrogen, 61-4801-82, 2338698; 1:100 dilution), 7-AAD (BD, 559925, 9031655; 1:40 dilution) or Ghost UV 450 Viability Dye (13-0868-T100, Tonbo Biosciences, D0868083018133, 1 µl ml−1) was used as viability dye. Flow cytometry was performed on an LSRFortessa instrument (BD Biosciences), FACS was performed on a SONY SH800S cell sorter and data were analyzed using FlowJo (TreeStar).
Single-cell RNA-seq
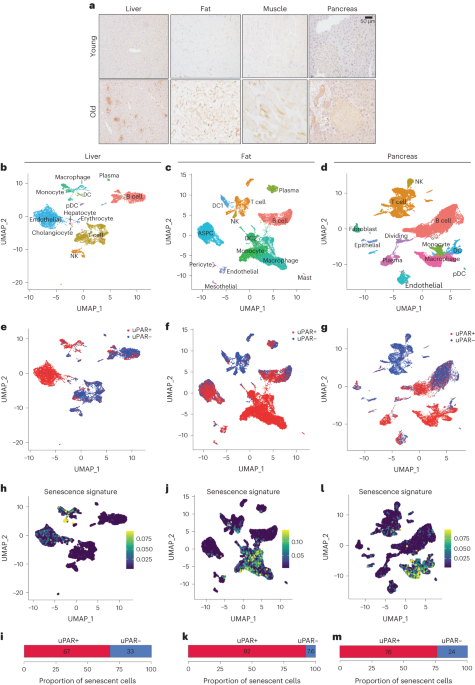
Sequencing data was demultiplexed, mapped, and processed into gene/cell expression matrices using 10x Genomics’ Cell Ranger software v7.1.0 (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger/). Gene expression reads were aligned to the mouse reference genome version gex-mm10-2020-A, available from the 10x Genomics website. We kept cells using the following parameters: ‘min.cells > 10, nFeature_RNA > 500, nCount_RNA > 2,500, percent.mt < 15’. Gene expression count data were normalized using SCTransform to regress out the percentage of mitochondrial RNA. The R package BBKNN was used to remove batch effects between mouse samples, and 0.5 was used as expression cutoff to define uPAR High cell populations. Clusters were identified using a resolution of 0.8, and cell types were annotated using R packages celldex, SingleR, Azimuth and custom gene sets20,21. Known markers for each cell type were plotted using the DotPlot function in Seurat. Senescence gene sets from refs. 17,24 were used to calculate signature scores using the AddModuleScore function in Seurat, and a signature score cutoff of 0.05 was used to define Senescence High cell populations. Differential expression analysis and functional annotations of gene sets were analyzed in the following way: Differential gene expression analysis was performed by comparing all the uPAR-positive versus uPAR-negative cells using RunPresto in Seurat, and the differentially expressed genes (DEGs) were determined by >1.5-fold change in gene expression with adjusted P value < 0.1. Pathway enrichment analysis was performed using the msigDB Hallmark gene sets using enrichR43. Significance of the tests was assessed using combined score, described as c = log(P) × z, where c is the combined score, P is Fisher’s exact test P value, z is the z-score for deviation from expected rank, and adjusted P values defined by enrichR. A lollipop plot was generated by plotting the top enriched/depleted log2(combined.score) on the x axis (directional), and size and color of the dots represents by −log10(adjusted P value).
Isolation, expansion and transduction of mouse T cells
B6.SJL-Ptrca/BoyAiTac mice (CD45.1 mice) were euthanized and spleens were collected. After tissue dissection and red blood cell lysis, primary mouse T cells were purified using the mouse Pan T cell Isolation Kit (Miltenyi Biotec). Purified T cells were cultured in RPMI-1640 (Invitrogen) supplemented with 10% FBS (HyClone), 10 mM HEPES (Invitrogen), 2 mM l-glutamine (Invitrogen), MEM non-essential amino acids 1× (Invitrogen), 55 µM β-mercaptoethanol, 1 mM sodium pyruvate (Invitrogen), 100 IU ml−1 recombinant human IL-2 (Proleukin; Novartis) and mouse anti-CD3/28 Dynabeads (Gibco) at a bead:cell ratio of 1:2. T cells were spinoculated with retroviral supernatant collected from Phoenix-ECO cells 24 h after initial T cell activation as described in refs. 44,45 and used for functional analysis 3–4 d later.
Genetic modification of T cells
The mouse SFG γ-retroviral m.uPAR-m28z plasmid has been described14. The mouse SFG γ-retroviral h.19-m28z plasmid14 was constructed by stepwise Gibson assembly (New England BioLabs) using the amino acid sequence for the scFv specific for human CD19 of the SFG-1928z backbone46 and cloned into the backbone of the SFG γ-retroviral m.uPAR-m28z plasmid14. In both constructs, the anti-mouse uPAR scFv or anti-human CD19 scFv is preceded by a mouse CD8A leader peptide and followed by the Myc-tag sequence (EQKLISEEDL), mouse CD28 transmembrane and intracellular domain and mouse CD3z intracellular domain44,45. Plasmids encoding the SFGγ retroviral vectors were used to transfect gpg29 fibroblasts (H29) to generate VSV-G pseudotyped retroviral supernatants, which were used to construct stable retrovirus-producing cell lines as described44,46.
Glucose tolerance testing
Blood samples from mice fasted for 8–12 h were collected at 0, 15, 30, 60 and 120 min after intraperitoneal injections of glucose (Sigma-Aldrich; 2 g per kg body weight for aging experiments and 1 g per kg body weight for HFD experiments). Insulin was measured from serum collected at the 0-min and 15-min time points. Concentrations were determined using the UltraSensitive Mouse Insulin ELISA kit (Crystal Chem, 90080).
Insulin tolerance testing
Blood samples from mice fasted for 4 h were collected at 0, 15, 30 and 60 min after intraperitoneal injections of insulin (Humulin R; Eli Lilly; 0.5 units per kg body weight).
Histological analysis
Tissues were fixed overnight in 10% formalin, embedded in paraffin and cut into 5-μm sections. Sections were subjected to H&E staining. Immunohistochemical staining was performed following standard protocols. The following antibodies were used: anti-mouse uPAR (AF534, R&D, DCL0521042; 1:50 dilution) and horse anti-goat IgG (30116; Vector Laboratories, ZH0526). Three fields per section were counted per sample with Fiji-ImageJ and averaged to quantify the percentage of uPAR-positive area per field. SA-β-gal staining was performed as previously described47 at a pH of 5.5 for mouse tissues. Specifically, fresh frozen tissue sections were fixed with 0.5% glutaraldehyde in PBS for 15 min, washed with PBS supplemented with 1 mM MgCl2 and stained for 5–8 h in PBS containing 1 mM MgCl2, 1 mg ml−1 X-gal, 5 mM potassium ferricyanide and 5 mM potassium ferrocyanide. Tissue sections were counterstained with eosin. Three fields per section were counted with ImageJ and averaged to quantify the percentage of SA-β-gal-positive area per field.
Immunofluorescence analysis
For the fluorescent SA-β-gal labeling, tissue slides were exposed to the C12RG substrate at 37 °C according to manufacturer’s instructions (ImaGene Red C12RG lacZ Gene Expression Kit, Molecular Probes, I2906)48,49. Subsequently, for immunofluorescence analysis, slides were fixed with 4% paraformaldehyde for 10 min at room temperature and regular immunofluorescence was performed following standard protocols and those previously described14. The following antibodies were used: anti-mouse uPAR uPAR (AF534,R&D, DCL0521042; 1:50 dilution) and anti-mouse F4/80 (Bio-Rad, CI:A3-1, 155529; 1:100 dilution). For quantification, five high-power fields per section were counted and averaged to quantify the percentage of SA-β-gal+, uPAR+ and F4/80+ per DAPI-positive cells. For colocalization analysis, Pearson coefficient was calculated using ImageJ.
Exercise capacity testing
Exercise capacity was assessed using a motorized treadmill (model 1050 EXER 3/6; Columbus Instruments). For 3 d before testing, mice were acclimatized to the treadmill (the mice walked on the treadmill at 10 m min−1 for 10 to 15 min per day). Following acclimatization, all mice underwent exercise capacity tests on consecutive days. Tests began with mice walking at 10 m min−1 with speed increased by 2 m min−1 every 2 min until exhaustion (mice could no longer achieve treadmill running speed despite repeated encouragement). The primary end points were time to exhaustion and maximum speed.
Blood measurements
Complete blood counts with differentials were performed using an automated hematology analyzer (IDEXX Procyte DX). For serum chemistry, blood was collected in tubes containing a serum separator. The tubes were then centrifuged, and the serum was obtained for analysis. Serum chemistry was performed by the LCP on a Beckman Coulter AU680 analyzer (Beckman Coulter Life Sciences). For cytokine analysis, plasma was collected and samples were processed and measured by Eve Technologies.
Pathology
Mice submitted for postmortem examination were euthanized by CO2 asphyxiation and cardiac exsanguination. Complete necropsies were performed at the Laboratory of Comparative Pathology (MSK, the Rockefeller University, and Weill Cornell Medicine). Representative sections were taken from all organ systems including heart, thymus, lungs, esophagus, trachea, thyroid glands, spleen, pancreas, liver, gallbladder, kidneys, adrenal glands, stomach, duodenum, jejunum, ileum, cecum, colon, lymph nodes (mesenteric and submandibular), salivary glands, skin (trunk and head), urinary bladder, epididymides, testes, prostate, seminal vesicles, uterus, cervix, vagina, ovaries, oviducts, spinal cord, vertebrae, sternum, femur, tibia, stifle joint, skeletal muscle, nerves, skull, nasal cavity, oral cavity, teeth, ears, eyes, pituitary gland and brain. Sections were fixed in 10% neutral-buffered formalin, processed in alcohol and xylene, embedded in paraffin, sectioned (5 μm thick) and stained with H&E. The skull, spinal column, sternum and hindlimb were decalcified in a formic acid and formaldehyde solution (Surgipath Decalcifier I, Leica Biosystems) before processing. H&E-stained tissue sections were evaluated by a board-certified veterinary pathologist (S.E.C.). Representative images were captured using a brightfield BX45 microscope with a DP26 camera and cellSens (version 1.18) Dimension software (Olympus America).
Statistical analysis
Data are presented as the mean ± s.e.m. Statistical analysis was performed by Student’s t-test or Mann–Whitney test using Prism v9.3.1 (GraphPad software). No statistical methods were used to predetermine sample size in the mouse studies, and no randomization method was used to allocate mice to experimental groups. Mouse conditions were observed by an operator who was blinded to the treatment groups in addition to the main investigator who was not blind to group allocation. Pathological analysis and exercise testing studies were performed in a blinded fashion. Data analysis was not performed in a blinded fashion. Data analysis was based on objectively measurable data (cell count, blood tests). No data were excluded except for histological assessment of HFD experiments, where we excluded OCT cassettes of samples containing adipose tissue or pancreas that were folded and presented a morphology that did not allow for successful slide generation; these were not further processed. Data distribution was assumed to be normal, but this was not formally tested. No adjustment for multiple comparisons was performed. The rationale for this was that to increase the rigor of select analyses, two control groups were compared to the experimental group, but it could have been biologically possible to just have one control group. Thus, for any given endpoint, there were two pairwise comparisons: the experiment group separately compared to each control. While two tests were evaluated, we only considered the analysis statistically significant if both tests had a P value less than 0.05. If only one of the two tests was significant, we did not claim the groups were significantly different; instead, we considered the analysis inconclusive and reported a trend. Viewing the analysis as significant only if both P values were less than 0.05 preserves the family-wise error rate at less than 0.05. Figures were prepared using BioRender.com for scientific illustrations in Figs. 3a and 7a,j, GraphPad Prism v9.3.1, and Microsoft Excel v16.77 and Illustrator CC 2022 (Adobe).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.