Mice
Husbandry
All procedures and animal husbandry were carried out in accordance with the UK Home Office under the Animals (Scientific Procedures) Act 1986, and the Crick Animal Welfare and Ethical Review Body, which is delivered as part of the Biological Research Facility Strategic Oversight Committee (BRF-SOC), under project license P609116C5. Mice were caged in individually ventilated cages, on a 12-h light–dark cycle, at ambient temperature and a humidity of 55 ± 10%, with food and water ad libitum. The maximum tumor size permitted under the project license P609116C5 is 1.5 cm in diameter, which was never exceeded.
WM tumor generation
To generate spontaneous non-recombined tumors as a source of biclonal tumors, Rosa26-CAG-lox-STOP-lox-MYC-ERT2/ Rosa26-mTmG/MMTV-Wnt1 mice were used. The transgenes used to generate the cross were on the following backgrounds: MMTV-Wnt1, FVB/N; Rosa26-mTmG, C57BL/6J and Rosa26-CAG-lox-STOP-lox-MYC-ERT2:Balb/c. The tumors arose spontaneously between 4 and 8 months of age. Once palpable, tumors were desensitized to tamoxifen by daily intraperitoneal (i.p.) injection (100 µl, 10 mg ml−1 tamoxifen (Sigma) in olive oil with 10% ethanol). At 1.5 mm3 tumors were excised, cut into small fragments and cryopreserved in FBS (Gibco, A5256701, lot 2575507) with 10% dimethylsulfoxide (DMSO). To generate secondary tumors, fragments were surgically implanted into the number four fat pad of female NOD/Scid mice ((NOD.CB17-Prkdcscid/NCrCrl) at the age of 6–8 weeks. Once tumors were palpable, mice were treated as above. At 1.5 mm3 tumors were excised and digested in 5 ml additive-free DMEM with 1 mg ml−1 Collagenase/Dispase (Roche) for 1 h under shaking. Tumor cells were suspended in high-glucose DMEM (Thermo Fisher Scientific, 11960044) with 2 mM glutamine and 10% FBS (Gibco, A5256701, lot 2575507). To generate WMhigh cells, Cre-expressing attenuated adenovirus (Ad5CMV-Cre, University of Iowa, VVC-U of Iowa-5) and Polybrene (8 μg ml−1), was added overnight. WMlow cells were not infected, but otherwise treated the same. Cells were washed extensively and eGFP- or tdTomato-positive cells were sorted with an Avalon sorter (BioRad). A total of 150,000 cells were then surgically implanted into the number four fat pad of 6–8-week-old NOD/Scid mice either as pure clonal populations or 1:1 mixtures. MYC-ERT2 was activated 3 d before collection by twice-daily i.p. injections of tamoxifen (100 µl, 10 mg ml−1 tamoxifen). On the day of collection, 3 h were left between the injection and further procedures, to have a fully active MYC-ERT2 construct. Tumors were collected at a size of 1.5 mm3. Mice were injected with BrdU (100 µl i.p., 10 mg ml−1 (Sigma)) 3 h before killing.
PDX tumor generation
The following previously described tumors were propagated via surgical fragment implant into 5-week-old female NSG mice subcutaneously26: HCI002, HCI009, STG143, STG195, STG335 and STG201; at the age of 5 weeks. Tumors took 2–10 months to form and were collected at a size of 1.5 mm3.
Diet modifications
A PA-free diet was formulated on the basis of a standard diet (composition is shown in Supplementary Table 2). PA absence was verified by LC–MS analysis. The control diet had the same composition as the PA-free diet, but with PA added. To avoid any interference with juvenile growth and puberty, diets were changed at 6 weeks of age. Diets were then introduced progressively over 4 d by increasing the amount of the PA-free/control diet and reducing the amounts of standard chow diet. In the meantime, tumor fragments were implanted into other Scid mice to grow WM tumors for in vitro recombination (WM only) and reimplantation or HCI002 tumors for reimplantation. Tumors were dissociated into a single-cell suspension and 750,000 (PDX) or 150,000 (WM tumors) cells were orthotopically implanted into the fourth inguinal fat pad of NOD/Scid mice. Mice were typically on a PA-free or control diet for 5 weeks before tumor implantation and remained on the diet until the tumors were collected. Dietary modification had no effect on mobility and responsiveness of animals nor their weight gain.
67NR cell-derived tumor generation
The 67NR control and SLC5A6 over-expressing cells (150,000 cells in 80% Matrigel) were transplanted into female Balbc/CJ mice at the age of 7 weeks.
Stable-isotope labeling in vivo
The following compounds were utilized for stable-isotope labeling: [13C6]glucose, [13C5]glutamine, [amide-15N]glutamine and calcium pantothenate ([13C3,15N]β-alanyl), all from Goss Scientific. We either performed bolus injections of the compounds to study acute consumption or infusions, to measure the steady-state label incorporation or repeated boluses over many days to study label accumulation (PA ([13C3,15N]β-alanyl)). Mice were not fasted before stable-isotope administration and all experiments were performed in the early afternoon.
Boluses were performed as previously described15. Stable-isotope-labeled compounds were diluted in saline and injected intravenously (i.v.) into the tail vein of the mice. [13C6]glucose was administered in a single bolus of 0.4 mg g−1 body weight in approximately 100 μl final volume. [13C5]glutamine has limited solubility and was thus administered in two boluses 15 min apart for a total amount of 0.34 mg g−1 body weight. In both cases, 15 min after the last injection, tissues and blood were collected. Tumors and control tissues were swiftly excised and either snap-frozen in liquid nitrogen or fixed. Blood was extracted by cardiac puncture and serum was separated for metabolite analyses.
In the case of PA ([13C3,15N]β-alanyl), when used in mice to label WM tumors for NanoSIMS analysis 100-μl boluses of 3 mg ml−1 PA were administered during five consecutive days. Tumor collection was performed 5 h after the last administration. Conversely, in the mice transplanted with 67NR SLC over-expressing and control cells, a single bolus of PA ([13C3,15N]β-alanyl) at 0.086 mg g−1 body weight was administered 15 min before tumor collection.
Infusions of isotopically labeled nutrients were performed under general anesthesia (isoflurane) using an Aladdin AL-1000 pump (World Precision Instruments) following previously published protocols42. For [13C6]glucose, mice received a bolus of 0.4 mg g−1 body weight, followed by a 0.012 mg g−1 body weight per minute infusion for 3 h. For glutamine-derived stable-isotope infusions, mice received a bolus of 0.187 mg g−1 body weight, followed by a 0.005 mg g−1 body weight per min infusion for 3 h. For co-infusion of [13C6]glucose and [amide-15N]glutamine, a solution of 40 mg ml−1 glutamine and 96 mg ml−1 glucose was prepared and mice received a bolus of 0.187 mg of glutamine and 0.442 mg glucose per gram of body weight. This was followed by an infusion of 0.005 mg glutamine and 0.012 mg glucose per gram of body weight per minute during 3 h. At the end of the infusion, tissue and blood were obtained as described for the boluses.
Where indicated, mice were injected with BrdU (100 µl i.p., 10 mg ml−1 (Sigma)) 3 h before killing.
Cell lines and culture conditions
The 4T1 and 67NR mouse mammary gland tumor cell lines were obtained from the Francis Crick Institute cell culture facility and were described previously. The 67NR–tet–cMYC–IRES–eGFP cells were described previously18,43.
The cell lines were authenticated using a standard protocol for identification of mouse cell lines using short tandem-repeat profiling. The profile is compared back to any available on commercial cell banks (such as ATCC and Cellosaurus). The species is confirmed by using a primer system, based on the Cytochrome C Oxidase Subunit 1 gene from mitochondria.
Cells were cultured in high-glucose DMEM (Thermo Fisher Scientific, 11960044) with 2 mM glutamine and 10% FBS (Gibco, A5256701, lot 2575507). Deviations are mentioned under the respective methods.
For in vitro PA uptake assays, cells were cultured in DMEM with 10 µM labeled PA ([13C3,15N]β-alanyl).
Human breast biopsies
The institutional review board approved collecting all samples for this study at Imperial College Healthcare National Health Service Trust (Imperial College Healthcare Tissue Bank HTA license no. 12275 and Tissue Bank sub-collection no. SUR-ZT-14-043). The REC no. (REC Wales approval) is 17/WA/0161. All patients provided their consent to use their samples in this study. All methods were performed according to institutional and ethical guidelines. Patients undergoing surgery were recruited and 12 tissue samples were taken from a range of subtypes, which were identified in the histopathology assessment (Extended Data Table 2). Data were only obtained on patients who had consented to the utilization of tissue for research. Tumors had to be of a macroscopic size ≥2 cm to allow for adequate research tissue without compromising the clinical diagnosis. Where feasible, tissue was provided from the center of the tumor from non-necrotic areas. Upon collection, all tissue samples were stored at −80 °C.
Metabolite extraction and analysis
Metabolite extraction
Snap-frozen tumors were ground with a liquid nitrogen-cooled mortar and pestle and subsequently lyophilized overnight in a FreeZone 4.5 Freeze Dry System (Labconco). Metabolites were extracted following published protocols44. Then, 5–10 mg of dried powder was weighed and extracted with 1.8 ml 2:1 chloroform:methanol. First, 0.6 ml methanol containing scyllo-inositol (10 nmol) and [13C5–15N]valine (5 µM final) as internal standards was added. This was followed by 1.2 ml chloroform containing margaric acid (C17:0, 10–20 μg). Samples were vortexed thoroughly and sonicated three times (8 min each) in a sonication bath at 4 °C. Samples were subsequently spun at 18,000g for 20 min and the supernatants were vacuum-dried in a rotational vacuum concentrator RVC 2–33 CD (Christ). Pellets were extracted with a methanol:water solution (2:1 v/v) as above and the two supernatants were combined and dried. Phase separation was performed with a 1:3:3 solution of chloroform:methanol:water. Polar (containing polar metabolites) and apolar (containing lipids) phases were stored separately at −80 °C until further processing. For the analysis of metabolites in blood, 5 μl serum was processed as above, without vortexing and sonication.
For in vitro assays, cells were plated in triplicate in six-well plates (250,000 cells per well) and cultured overnight before changing the medium to the indicated experimental conditions. At collection, cells were washed with PBS and snap-frozen. Subsequently, cells were scraped in 0.75 ml methanol, 0.25 ml chloroform and 0.25 ml water containing 1 nmol scyllo-inositol as an internal standard. Samples were vortexed, sonicated for 3 × 8-min bursts at 4 °C and incubated overnight at 4 °C. Samples were subsequently pelleted by centrifugation at 20,000g for 20 min at 4 °C. Phase separation was performed with a 1:3:3 solution of chloroform:methanol:water through the addition of 0.25 ml water to the supernatant. Polar and apolar phases were separated by centrifugation at 20,000g for 5 min at 4 °C. The polar phase was vacuum-dried in a rotational vacuum concentrator RVC 2–33 CD (Christ) for GC–MS analysis as detailed below.
GC–MS analysis
GC–MS analysis was performed following published protocols15. Part of the polar fraction was dried and washed twice with methanol. The first wash contained l-Nor-leucine (1.6 nmol per sample) as a run standard. For derivatization, metabolites were incubated with methoximation (Sigma, 20 µl, 20 mg ml−1 in pyridine) overnight followed by trimethylsilylation (20 µl N,O-bis(trimethylsilyl)trifluoroacetamide reagent (BSTFA) containing 1% trimethylchlorosilane (TMCS), Thermo Fisher). Samples were then analyzed using an Agilent 7890A-5975C GC–MS system. Splitless injection (injection temperature 270 °C) onto a 30 m + 10 m × 0.25 mm DB-5MS + DG column (Agilent J&W) was used, using helium as the carrier gas, in electron ionization mode. The initial oven temperature was 70 °C (2 min), followed by temperature gradients to 295 °C at 12.5 °C per min and then to 320 °C at 25 °C per min (held for 3 min). Under these conditions, glutamine and glutamic acid can spontaneously cyclize to pyroglutamic acid. The latter was thus used to assess glutamine levels in the tissue samples or plasma extracts, due to its preponderant abundance in blood. Metabolites were identified based on a mix of authentic standards, using the MassHunter software (Agilent, v.10.0.368). Label incorporation was calculated by subtracting the natural abundance of stable isotopes from the observed amounts15,45. Briefly, The level of enrichment of individual isotopologs (m+x) of metabolites was estimated as the percentage of the metabolite pool containing x 13C atoms after correction for natural abundance:
$${\rm{Enrichment}}\; {\rm{of}}\; m+{xx}=\frac{{\rm{Cm}}+x}{\varSigma {\rm{Cm}}+0+{\rm{Cm}}+1\ldots +{\rm{Cm}}+i}\times 100 \%$$
The percentage carbons enriched for a metabolite with i isotopologs was calculated by:
$$13{\rm{Cmet}}=\frac{m=1}{i}+2\frac{m+2}{i}\ldots +i\frac{m+1}{i}$$
LC–MS analysis
LC–MS analysis of the polar extracts was carried out following published protocols46. Samples were injected into a Dionex UltiMate LC system (Thermo Scientific) with a ZIC-pHILIC (150 mm × 4.6 mm, 5-μm particle) column (Merck Sequant). Solvent B was acetonitrile (Optima HPLC grade, Sigma-Aldrich) and solvent A was 20 mM ammonium carbonate in water (Optima HPLC grade, Sigma-Aldrich). A 15-min elution gradient of 80% solvent A to 20% solvent B was used, followed by a 5 min wash of 95:5 solvent A to solvent B and 5 min re-equilibration. Other parameters were a flow rate of 300 µl min−1; column temperature of 25 °C; injection volume of 10 µl; and autosampler temperature of 4 °C. MS was performed with positive/negative polarity switching using an Q Exactive Orbitrap (Thermo Scientific) with a HESI II probe. MS parameters were spray voltage of 3.5 kV and 3.2 kV for positive and negative modes, respectively; probe temperature of 320 °C; sheath and auxiliary gases were 30 and 5 AU, respectively; full scan range was 70 to 1050 m/z with settings of auto gain control (AGC) target and resolution as Balanced and High (3 × 106 and 70,000), respectively. Data were recorded using Xcalibur software (Thermo Scientific), v.3.0.63. Mass calibration was performed for both ESI polarities before analysis using the standard Thermo Scientific Calmix solution. To enhance calibration stability, lock-mass correction was also applied to each analytical run using ubiquitous low-mass contaminants. Quality control samples were prepared by pooling equal volumes of each sample and analyzed throughout the run to provide a measurement of the stability and performance of the system. To confirm the identification of important features, some quality control samples were run in data-dependent top-N (ddMS2-top-N) mode, with acquisition parameters as follows: resolution of 17,500; AGC target under 2 × 105; isolation window of m/z 0.4; and stepped collision energy 10, 20 and 30 in high-energy collisional dissociation mode. Qualitative and quantitative analysis was performed using Free Style v.1.5 and Tracefinder v.4.1 software (Thermo Scientific) according to the manufacturer’s workflows. For putative annotation, a CEU Mass Mediator tool was employed47.
LC–MS analysis of the apolar extracts was carried out following published protocols48. Lipids were separated by injecting 10-μl aliquots onto a 2.1 × 100 mm, 1.8-μm C18 Zorbax Eclipse plus column (Agilent) using a Dionex UltiMate 3000 LC system (Thermo Scientific). Solvent A was 10 mM ammonium formate in water (Optima HPLC grade, Fisher Chemical) and solvent B was water:acetonitrile:isopropanol, 5:20:75 (v/v/v) with 10 mM ammonium formate (Optima HPLC grade, Fisher Chemical). A 20-min elution gradient of 45% to 100% solvent B was used, followed by a 5 min wash of 100% solvent B and 3 min re-equilibration. Other parameters were a flow rate of 600 μl min−1; a column temperature of 60 °C; and an autosampler temperature of 10 °C. MS was performed with positive/negative polarity switching using a Q Exactive Orbitrap (Thermo Scientific) with a HESI II probe. MS parameters were a spray voltage of 3.5 kV and 2.5 kV for positive and negative modes, respectively; a probe temperature of 275 °C; sheath and auxiliary gases were 55 and 15 arbitrary units, respectively; full scan range was 150 to 2,000 m/z with settings of AGC target and resolution as Balanced and High (3 × 106 and 70,000), respectively. Data were recorded using Xcalibur software (Thermo Scientific), v.3.0.63. Mass calibration was performed for both ESI polarities before analysis using the standard Thermo Scientific Calmix solution. To enhance calibration stability, lock-mass correction was also applied to each analytical run using ubiquitous low-mass contaminants. To confirm the identification of significant features, pooled quality control samples were run in data-dependent top-N (ddMS2-top-N) mode, with acquisition parameters of mass resolution of 17,500; AGC target under 2 × 105; isolation window of m/z 0.4; and stepped collision energy of 10, 20 and 30 in high-energy collisional dissociation mode. Qualitative and quantitative analyses were performed using Free Style v.1.5 (Thermo Scientific), Progenesis QI v.2.4.6911.27652 (Nonlinear Dynamics) and LipidMatch v.2.02 (Innovative Omics)49. Radial plot representations were performed in R (v.3.6.2) using the R package volcano3D v.2.08 (ref. 50).
For the detection of CoA-SH and acetyl-CoA we used an Agilent 1200 LC system equipped with an Agilent Poroshell 120 EC-C18, 2.7 μm, 4.6 × 50 mm column was used. Then, 10 μl cleared samples were injected on the column. Mobile phase A was 40 mM ammonium formate at pH 6.8 and mobile phase B was LC–MS grade acetonitrile. Metabolites were separated at a flow rate 0.5 ml min−1, using the following elution conditions: 0–1 min, 2% B; 1–10 min, 2–98% B; and 10–12 min, 98% B. An Agilent accurate mass 6230 time-of-flight apparatus was employed. Dynamic mass axis calibration was achieved by continuous infusion of a reference mass solution using an isocratic pump connected to a dual Agilent Jet Stream ESI source, operated in the positive-ion mode. ESI capillary, nozzle and fragmentor voltages were set at 3,000 V, 2,000 V and 110 V, respectively. The nebulizer pressure was set at 40 psi and the nitrogen drying gas flow rate was set at 10 l min−1. The drying gas temperature was maintained at 200 °C. The sheath gas temperature and flow rate were 350 °C and 11 l min−1. The MS acquisition rate was 1.0 spectra s−1 and m/z data ranging from 50–1,200 were stored. The instrument routinely enabled accurate mass spectral measurements with an error of <5 ppm. Data were collected in the 2 GHz (extended dynamic range) mode and stored in centroid format.
Western blot
Western blots were run using standard protocols for non-native protein detection. The following antibodies were used: MYC: AbCam, ab32072, 1:1,000 dilution; SLC5A6: Proteintech 26407-1-AP, 1:1,000 dilution; actin: Merck A3854 (HRP coupled), 1:25,000 dilution; cleaved caspase 3, Cell Signaling cat. no. 9664, 1:1,000 dilution; ATF4: AbCam, ab23760, 1:1,000 dilution; PDHE1: AbCam ab110330, 1:1,000 dilution; p70S6K: Cell Signaling, cat. no. 9202, 1:1,000 dilution; p-p70S6K: Cell Signaling, cat. no. 9234, 1:1,000; p-S6: Cell Signaling, cat. no. 4834s, 1:1,000 dilution; S6k: Cell Signaling, cat. no. 2217, 1:1,000 dilution; HK2: Cell Signaling, cat. no. 2867S, 1:1,000 dilution; AcSL1: Cell Signaling, cat. no. 4047, 1:1,000 dilution; p-PERK (Thr980): Cell Signaling, cat. no. 3179, 1:1,000 dilution; PERK: Cell Signaling, cat. no. 3192, 1:1,000 dilution; p-eIF2ɑ (Ser51): Cell Signaling, cat. no. 3398, 1:1,000 dilution; eIF2ɑ: Cell Signaling, cat. no. 5324, 1:1,000 dilution. The secondary antibodies were anti-rabbit-HRP, GE Healthcare, cat. no. NA934-1ML, 1:7,500 dilution; and anti-mouse-HRP, Invitrogen, cat. no. 62-6520, 1:7,500 dilution.
RNA extraction and quantitative PCR
The 67NR-tet-cMYC cells were plated at 1.5 × 105 cells in a six-well plate and cultured overnight before induction of MYC with doxycycline for 24 h. Cells were collected in TRIzol (Thermo Fisher). Similarly, 3 mg tumor tissue was homogenized in TRIzol. Total cellular RNA was extracted according to the manufacturer’s instructions. Subsequently, RNA was treated with DNase (Thermo Fisher, EN0525) and 1 µg RNA was incubated with 0.5 µM random primers. Complementary DNA was generated using Superscript III Reverse Transcriptase (Invitrogen, 18080044) and treatment with RNase OUT (Invitrogen, 10777019) according to the manufacturer’s instructions. Quantitative PCR reactions were performed in a ViiA 7 Real-Time PCR System (Thermo Fisher). PCR reactions were performed using Applied Biosystems Power SYBR Green PCR Master Mix (Thermo Fisher) in 10-µl reactions containing 5 pmol forward and reverse primer at 200 ng of cDNA. PCR conditions were an initial denaturation and polymerase activation for 10 min at 95 °C, followed by 40 cycles of 15 s and 95 °C and 60 s at 60 °C. These amplification cycles were followed by a melt-curve denaturation. Fold change and absolute abundance were determined using the 2ΔΔCT and 2ΔCT methods, respectively.
Primer sequences were designed using NCBI-PrimerBLAST:
c-MYC: (F) 5′-CCTTCTCTCCGTCCTCGGAT-3′, (R) 5′-TCTTGTTCCTCCTCAGAGTCG-3′; Slc5a6: (F) 5′-TTCACTGGCAACTGTCACGA-3′, (R) 5′-AGATACTGAGTGCTGCCTGG-3′; Pank1: (F) 5′-AAGAACAGGCCGCCATTCC-3′, (R) 5′-CTGCCGTGATATCCTTCGT-3′; Pank2: (F) 5′-TCACAGGCACCAGTCTTGGA-3′, (R) 5′-CCTGGCAAGCCAAACCTCT-3′;; Ppcdc: (F) 5′-CGTGCGTGTTGAGGTCATAG-3′, (R) 5′- GCCTGGGTCTAGAATCTGTCA-3′; Aasdhppt: (F) 5′-AAAGGGAAAGCCGGTTCTTG-3′, (R) 5′-ATTGAACCACGACCTGGAAA-3′; Slc1a5: (F) 5′-GCCTTCCGCTCTTTTGCTAC-3′, (R) 5′-GACGATAGCGAAGACCACCA-3′,
SLC5A6 exogenous: (F)TGGGCTGCTGTTACTTCCTG, (R) CACCCTCACGGTCTTGTTGA,
The sequence for primers for Slc38a1 were obtained from Bian et al.51: (F) 5′-TACCAGAGCACAGGCGACATTC-3′, (R) 5′-ATGGCGGCACAGGTGGAACTTT-3′.
Cloning
Using Gateway cloning (Invitrogen, Thermo Fisher), SLC5A6 (pDONR221_SLC5A6 was a gift from the RESOLUTE Consortium and G. Superti-Furga (Addgene plasmid #132194)) was inserted into the retroviral plasmid, pBabe-Puro (pBABE-Puro-gateway was a gift from M. Meyerson (Addgene plasmid #51070)).
Retroviral gene transfer
Phoenix-AMPHO cells were seeded at 1.0 × 106 cells in 6-cm plates and cultured overnight. Cells were subsequently transfected with 2 µg pBabe-SLC5A6 plasmid using PEI. After 16 h, the medium was replaced with 5 ml DMEM + 10% FBS and cells were cultured for an additional 48 h before collection of virus-containing supernatant. The supernatant was centrifuged at 1,000g for 5 min at 4 °C and filtered through a 0.45-µm syringe filter. Before transduction, 67NR cells were grown to 80% confluency and treated with 8 µg ml−1 Polybrene before infection with 0.5 ml virus-containing medium. After 24 h of transduction, cells were replated with 3 mg ml−1 puromycin for 72 h and maintained in the culture medium at 1 μg ml−1.
Growth curves
Cells were washed with PBS before seeding at indicated confluency in high-glucose DMEM (custom made at the Francis Crick Institute) with 10% dialyzed FBS52 with or without 10 µM d-calcium pantothenate (Sigma, P5155) in 48-well plates. Cells cultured in the absence of pantothenate were cultured with or without Coenzyme A Trilithium Salt (EMD Millipore, 234101) at the indicated concentrations. Cells were imaged recurrently with the Incucyte S3 Live Cell Analysis System and confluency was measured using the Incucyte S3 Software (Sartorius) v.2021C.
Sample preparation and DEFFI-MSI
Snap-frozen tumors were mounted onto cryosectioning chucks with a freezing drop of ice. The tumors were sectioned individually into 10-μm slices at −21 °C and thaw-mounted onto SuperFrost glass slides of 75 mm × 25 mm (Thermo Fisher Scientific). Sections were dried with a flow of nitrogen, placed in slide boxes and vacuum packed. They were stored at −80 °C until they were used for analysis. One set of PDXs was instead embedded in hydroxypropyl-methylcellulose (40–60 cP, 2% in H2O) and polyvinylpyrrolidone (average molecular weight 360,000) 7.5 and 2.5 g, respectively in 100 ml H2O and cryosectioned53. The downstream processing was as described above.
For human breast cancer biopsies each sample was cryosectioned into 10-μm thick parallel sections using a Cryostat Leica CM 1950 (Leica) set to −27 °C in the chamber and −25 °C in the sample holder. Tissue sections were put onto SuperFrost plus glass slides (Thermo Fisher Scientific). The slides were vacuum packed and stored at −80 °C until DEFFI-MSI analysis.
Imaging was carried out on the DESI imaging source (Prosolia) consisting of a two-dimensional sample holder moving stage coupled to a XEVO G2-XS QToF (Waters CorporationA), with the ion block temperature set at 150 °C. The DESI sprayer was converted to a DEFFI sprayer according to Wu et al. 20 by pulling the solvent capillary inwards20. A custom-built inlet capillary was heated up to 500 °C to assist the desolvation of secondary droplets. All images were acquired using HDImaging v.1.4 software in combination with MassLynx v.4.1 (Waters Corporation). Imaging parameters were set as follows: N2 gas pressure at 5 bar; 95:5 v/v methanol/water solvent was delivered by a nanoAcquity binary solvent manager (Waters Corporation) set at a flow rate of 1.5 µl min−1; sprayer voltage at 4.5 kV; sprayer angle of 75°; sprayer to surface distance of 2 mm; sprayer to inlet capillary distance of 1 mm; sprayer inlet capillary collection angle of 10°; emitter orifice distance of 150 µm; and orifice diameter of 200 µm. MS parameters were set as follows: m/z 50–1,000, one scan per second, horizontal acquisition speed at 100 μm s−1 with a lateral resolution of 100 μm, acquired in negative sensitivity MS mode. PA standard (100 ppm in 50:50 (v/v) methanol:water) was spiked onto tissue and left at room temperature for 15 min to dry. DEFFI-MS/MS imaging was then performed in negative sensitivity mode (precursor ion m/z of 218.10; collision energy of 12 v; one scan per second; and 100 µm × 100 µm pixel size). Once acquisition was finished, all tissue sections were stored at −80 °C before staining.
After DEFFI acquisitions, samples were rehydrated. For WM tumors, the slides were immediately cover-slipped and fluorescence was imaged on an Olympus VC120 slide scanner. PDX samples were also rehydrated and fixed for 10 min in 4% PFA. Staining was performed following standard IHC protocols.
DEFFI image analysis
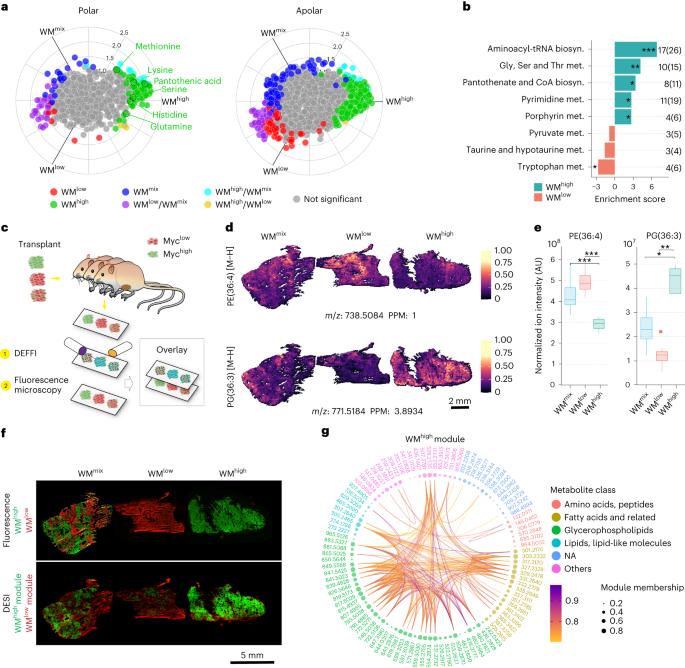
DEFFI-MSI peaks used for the statistical modeling were extracted from the raw spectra. As a first step, peaks were centroided using the method described by Inglese et al.22. To include possible shoulder peaks, we selected those with a prominence greater than five. Peaks prominences were estimated using the ‘find_peaks’ function available in the Scipy package v.1.6.3 for Python54. A first intra-run peak matching was performed using MALDIquant v.1.19.3 (ref. 55) relaxed method with a search window of 50 ppm. This allowed SPUTNIK spatial filtering to remove signals that were detected outside the tissue or were associated with scattered images (number of eight-connected pixels <9) (ref. 56). Subsequently to the peak filtering, all non-tissue pixels were discarded. The filtered tissue peaks were mapped back to their original raw m/z values and mass recalibrated using the method described by Inglese et al.22. The recalibrated intra-run peaks were therefore matched using MALDIquant v.1.19.3 relaxed method. Matching of tissue peaks was performed with a search window of 50 ppm. If more than one peak was found in the search windows in the same spectrum, only the one with the highest intensity was retrieved. After this step, each run was assigned a set of common masses. To match the peak masses among different runs, for each DEFFI-MSI run r∈R, a representative list of peaks was calculated as the set of ordered pairs Xr = {(m/zp,r, Yp,r): p∈Pr }, where \({Y}_{p,r}={\sum }_{i=1}^{N}{y}_{i,p,r}/N\) is the arithmetic mean of the pixel-wise intensities yi,r of the peak p with mass-to-charge ratio m/zp,r and Pr is the set of common m/z values of the run r. The m/z values of the representative peaks lists of all R runs X′ = {X1, X2,…,XR} were matched together using the MALDIquant v.1.19.3 relaxed method with a tolerance of 20 ppm to generate a set P′ of inter-run common m/z values. Thus, this set of masses was common to all runs. The set of inter-run common m/z values was therefore filtered using a consensus approach. We removed all peaks p′∈P′ if the corresponding mean intensity Yp′,r was equal to zero in at least one run r∈R. Final intensity matrices were median-scaling normalized (using median of non-zero intensities) and batch-effect corrected using ComBat (from SVA package v.3.34.0)57, with the batch equal to the acquisition run.
WGCNA v.1.70-3 (ref. 58) was employed to determine a consensus metabolic network following a similar approach described by Inglese et al.21. Only spectra corresponding to WMhigh and WMlow tumors, not WMmix were used to estimate the network. Signed hybrid58 adjacencies, corresponding to the only positive Pearson’s correlations between all pairs of features, were calculated from the individual runs, using a consensus soft power equal to the smallest value corresponding to an R2 > 0.85 across all runs. Tested values for the soft power ranged from 1 to 20. The adjacencies were subsequently normalized using the ‘single quantile’ method available in the WGCNA v.1.70-3 package for R, with the reference quantile set equal to 0.95 and combined into a consensus adjacency. Each element of the consensus adjacency matrix corresponded to the minimum adjacency value across the runs.
The consensus topological overlap matrix59,60, estimated from the consensus adjacency, was used to determine the network modules through hierarchical clustering (average linkage).
An initial set of clusters were determined using the dynamicTreeCut algorithm61, with the ‘hybrid’ algorithm and a smallest cluster size equal to 20. Module eigenmetabolites (MEs) were calculated from each module as the first principal-component scores of the merged runs spectra, using the only module features. The scores sign was reversed if Pearson’s correlation with the average image (calculated assigning to each pixel the mean intensity of their peaks) was negative. Modules with MEs correlated (Pearson’s correlation) more than 0.85 were considered identical and merged into a single module. Final MEs were recalculated from the merged modules. Association between modules and MYC was estimated using a linear regression model. First, for each module, the WMhigh and WMlow values of the ME were binarized into ‘low-ME’ and ‘high-ME’ using the inter-run median value as threshold. Then, the proportion of ‘high-ME’ was calculated within each tissue section and used as the dependent variable of the regression model, whereas the binary tissue MYC condition of each pixel was used as independent variable. Because of its proportion nature, the dependent variable was modeled as a β-distributed (link = ‘logit’)62,63. ME models with a significantly different from zero slope (partial Wald test P < 0.05, Benjamini–Hochberg correction) were considered statistically associated to MYC. Finally, ME values for the WMmix spectra were calculated projecting their module peak intensities on the corresponding loadings estimated from the WMhigh and WMlow spectra.
All DEFFI single-ion images were maximal intensity normalized for individual ions within each run and scales were adjusted to values of 0–1 with the color palette adjusted with thresholding intensity at the 99.9th percentile to avoid artifacts caused by outliers, unless specified.
Dendrogram for ion colocalization of labeled metabolites
Mass-to-charge ratios (m/z) of selected metabolites were searched in the list of inter-run matched common m/z values. Theoretical m/z values were calculated from the deprotonated mass adding k times the δ m/z corresponding to the mass difference between 13C and 12C (ΔC = 1.003355 m/z). The value of k varied from zero to five, where k > 0 represented the isotopic forms. Features within an error of 10 ppm from the theoretical m/z value were considered as candidate matches. In the case of multiple candidates, the one corresponding to the highest mean intensity across all pixels was selected.
Matched features intensities corresponding to isotopes (k > 0) were adjusted for the natural isotopic abundance to estimate the intensity corresponding to the only labeled metabolites. The natural isotopic abundance was estimated from the observed raw monoisotopic intensities, with the probability of observing k 13C atoms, given N carbon atoms, modeled as binomially distributed, Pr(X = k) = Binom(k, N, P), where P = 0.01109 represents the probability of observing natural 13C isotopes. Pixel intensities corresponding to the labeled 13C component were therefore calculated subtracting the estimated natural 13C intensity from the raw intensity of the same pixel. Negative intensities could result from measurement errors. In these cases, we set the 13C intensity to zero, following the assumption that either no labeled molecules were detected or their abundance fell below the limit of detection of the instrument.
All three runs were included in the calculation. Intra-run Pearson’s correlations were estimated between the corrected intensities of the matched features. Subsequently, an inter-run consensus correlation matrix was calculated by taking the minimum values among the runs. Only WMmix pixels were used for the calculation of the correlations.
The null hypothesis that consensus correlations between ion pairs is zero was tested using a permutation test. In each of 10,000 permutations, consensus correlations were calculated after shuffling the pixel intensities of the individual ion images. P values were calculated as \(({\sum }_{i}{I}_{i}+1)/10,001\), where Ii is the indicator function, which is equal to 1 if the absolute permuted correlation is greater or equal than the original absolute correlation, 0 otherwise. P values of all pairwise correlations were adjusted using the Benjamini–Hochberg method.
A hierarchical clustering, with average linkage, was employed to partition the matched metabolites. One minus the consensus correlation was used as a distance measure. Clusters were identified by cutting the dendrogram at the level corresponding to two clusters.
Automated segmentation and co-registration of PDX samples and human biopsies
Levels of MYC were identified by IHC (see below) in PDX samples and human biopsies. The IHC signal was deconvoluted using the inbuilt DAB staining deconvolution algorithm of the QuPath software package. The pixels devoid of any DAB staining from the deconvoluted IHC images were then removed by thresholding the green image channel at an intensity of 230. The remaining pixels were subsequently assigned as MYC positive. The image was then re-binned by a factor of 16 by summing the total number of MYC-positive pixels in any given 16 × 16 area using the MATLAB function ‘blockproc’ (Mathworks, image-processing toolbox). The resulting images (the MYC percentage proportions figures) were then clustered using the k-means clustering algorithm using Euclidean distance and k = 2 to differentiate regions that are high in MYC stain versus low. The binary image of the cluster that was highest in MYC signal versus those with low and no MYC was then extracted and registered to the MSI image. Areas of overt necrosis, staining and processing artifacts were excluded from the analysis by drawing masks. The masks were applied to the single-ion images for PA from MSI analysis as well as to the deconvoluted MYC staining before binarization, and within each tumor the average difference of mean levels of PA between regions of high and low MYC was assessed by a linear mixed-effects model fitted with the ‘glmmTMB‘ v.1.1.4 package for R (random intercepts were considered for run and tissue ID). We then plotted the observed proportions connected by a line per sample, with a black dot representing the predicted mean of the two groups. The error bars represent the confidence interval of the predictions for the two groups (Supplementary Fig. 1).
Immunohistochemistry/immunofluorescence
Immunofluorescence staining was performed against BrDU, eGFP and Tomato using the following antibodies. BrDU (BD, 347580, 1:100 dilution); eGFP *(Abcam, ab6683, 1:100 dilution); and RFP (Rockland, 600-401-379, 1:500 dilution). Secondary antibodies were Alexa Fluor-647 donkey anti-mouse, A-31571, Alexa Fluor-555 donkey anti-rabbit, A-31572, Alexa Fluor-488 donkey anti-goat, A-11055, all at 1:500 dilution. Slides were blocked for 10 min in 4% PFA at room temperature and rehydrated in PBS–Tween (0.1%) for 10 min. Slides were then transferred into a citric buffer (10 mM at pH 6.0) and boiled in a microwave for 15 min. Slides were cooled under running tap water and blocking was performed in PBS–Tween (0.1%) with 3% BSA for 1 h at room temperature. Primary antibodies were incubated overnight, whereas secondary antibodies were incubated for 1 h at room temperature. Nuclei were stained with Hoechst (0.1 μg ml−1, Sigma). To assess proliferation in WM tumors, three visual fields per sample were segmented by clones (WMhigh and WMlow) and total nuclei as well as BrdU-positive nuclei were quantified using the cell counting plugin in ImageJ. To assess proliferations in PA-deprived and control HCI002 PDXs, BrdU staining was carried out as above, slides were scanned with an Olympus VC120 slide scanner and nuclei as well as BrdU-positive cells were quantified on the whole slide using the Qpath cell-counting feature. IHC staining for c-MYC was carried out on frozen sections post-DEFFI for PDX samples and on the consecutive slice for human biopsies. The secondary antibody was a biotinylated goat anti-rabbit (BA-1000, Vector, 1:250 dilution) for 45 min at room temperature. The ABC kit (PK-6100 from Vector) was incubated for 30 min and DAB for 10 min.
NanoSIMS
Electron microscopy embedding
For NanoSIMS analysis, WMmix tumors were grown as described above. At 3 h before collection, BrdU (100 μl, 10 mg ml−1) was administered to the mice i.p. Either boluses of stable-isotope labeled calcium pantothenate ([13C3,15N]β-alanyl) or a co-infusion of [13C6]glucose and [amide-15N]glutamine were administered to the mice as described above. Once removed tumors were fixed overnight in freshly prepared 4% paraformaldehyde in 0.1 M phosphate buffer (PB) at pH 7.4 and stored at 4 °C. Following initial fixation, samples were embedded in 2% low-melting-point agarose (A4018-50G, Sigma-Aldrich) in 0.1 M PB and 150-μm sections were collected using a vibrating knife ultramicrotome (VT1200S, Leica), using a speed of 1 mm s−1 and an amplitude of 0.75 mm. Excess agarose was removed from the sections and tissue was stored in a 24-well plate in 0.1 M PB at 4 °C.
Sections were then transferred onto a glass slide, cover-slipped in PB and the whole section imaged in a single plain with a confocal microscope (Leica, Falcon SP7) at ×100 magnification (objective HCPL APO CS2 ×10, NA 0.4). Regions of interest (ROIs) were chosen and a z-stack of approximately 70-μm depth at ×100 magnification was imaged over an area of approximately 1.5 mm2.
Imaged sections were then removed from the glass slide and embedded using a protocol adapted from the NCMIR method64. Sections were post-fixed in 4% paraformaldehyde/2.5% glutaraldehyde in 0.1 M PB at pH 7.4 for 1 h at room temperature. Samples were then washed in 0.1 M PB (5 × 3 min) before being post-fixed in 2% reduced osmium (2% osmium tetroxide/1.5% potassium ferricyanide) at 4 °C for 1 h. Sections were washed (5 × 3 min in dH2O), stained in 1% thiocarbohydrazide for 20 min at room temperature, washed again (5 × 3 min in dH2O) and stained in 2% osmium tetroxide for 30 min at room temperature. Finally, samples were washed (5 × 3 min in dH2O) before being left overnight in 1% uranyl acetate at 4 °C. The following day sections were washed (5 × 3 min in dH2O) and then stained en bloc with lead aspartate (pH 5.5) for 30 min at 60 °C. After a final wash (5 × 3 min in dH2O), sections were dehydrated using a graded series of alcohol (20%, 50%, 75%, 90%, 100% × 2, 20 min each) followed by infiltration with Durcupan resin (44610-1EA, Sigma-Aldrich) (1:1 resin:ethanol overnight and 100% resin for 24 h). Sections were then flat embedded using Aclar (L4458, Agar Scientific) and polymerized at 60 °C for 48 h.
Targeted single-section large area montaging
Polymerized sections were removed from the Aclar and blocks were prepared from the ROI. The ROI was identified by aligning the overview ×100 confocal image of the whole section to an overview image of the now-embedded section, acquired using a stereo microscope (MC205C stereo, DMC 4500 Camera, Leica Microsystems) in Photoshop (Adobe). The identified area was then removed from the resin with a small bit of excess on each side using a razor blade and the excised block was mounted on a metal pin (10-006002-50, Labtech) using silver epoxy (604057, CW2400 adhesive, Farnell), which was polymerized at 60 °C for 1 h65. The sample was removed from the oven and any excess resin and silver epoxy was trimmed using a glass knife on an ultramicrotome (EM UC7, Leica Microsystems). The resin block was shaped into a square with one corner removed so that it was asymmetric, to aid in orientation of the block in the serial block-face (SBF) scanning electron microscope (SEM). Finally, the block face was trimmed until the tissue was reached. The block was then sputter coated with 10 nm platinum (Q150S, Quorum Technologies) and loaded into a 3View2XP (Gatan) attached to a Sigma VP SEM (Zeiss) with focal charge compensation (Zeiss). The SBF SEM was used as a ‘smart trimming’ tool, allowing visual assessment of the tissue structure during cutting until the EM images of the block face, collected using a BSE detector (3View detector, Gatan), matched with the structures imaged in the ×100 confocal z-stack. When the z-plane containing the ROI had been reached, the sample was removed from the SBF SEM, the block was re-trimmed using a glass knife to a sub-area of the ROI approximately 400 μm × 200 μm and 200-nm sections were cut from the block face using an ultramicrotome (EM UC7, Leica Microsystems) and a 6-mm histo diamond knife (DiATOME). Sections were collected onto silicon wafers (G3390, Agar Scientific) using an eyelash and dried on a hotplate at 70 °C for 10 min. The silicon wafers were then mounted onto SEM stubs (10-002012-100, Labtech Electron Microscopy) using adhesive carbon tabs (15-000412, Labtech Electron Microscopy) and loaded into a Quanta FEG 250 SEM (Thermo Fisher Scientific). Tiled images of the whole section were collected using Maps software (v.1.1.8.603, Thermo Fisher Scientific) using a low-voltage high-contrast backscattered electron detector (vCD, Thermo Fisher Scientific). Images were collected using a voltage of 2.5 kV, a spot size of 3, a dwell time of 5 ms, a working distance of 6 mm and a pixel resolution of 10 nm. Individual images from the tiled sequence were exported to TIFF files and aligned into a single image using the TrackEM2 plugin in Fiji66. Both the exported single image and the corresponding resin section on a silicon wafer were then sent to NanoSIMS for targeted analysis, where they were loaded into the NanoSIMS (Cameca NanoSIMS 50L, Cameca/Ametek). Before analysis, the pulse height distributions of the electron multiplier detectors were analyzed and their voltage gains and thresholds adjusted if necessary. They were then further examined by measuring the C− and CN− count rate on adjacent detectors used to measure the 13C/12C and 12C15N/12C14N isotope ratios. This step is imperative to measure accurate isotope ratios. The NanoSIMS 50L has seven detectors which were then moved to the appropriate radii in the magnetic sector with fixed magnetic field to measure the following masses: 12C, 13C, 12C14N, 12C15N, 31P, 79Br and 81Br. Images were typically acquired at 50 μm × 50 μm field width or 25 μm × 25 μm with a 300-μm D1 aperture (D1-2) to provide a mosaic correlating with regions previously analyzed by fluorescence and scanning EM. Nono-SIMS images were acquired using Cameca NanoSIMS N550L software v.4.4. and were processed and quantitative data extracted using the OpenMIMS plugin for ImageJ.
The acquisition modes were overlaid using the BigWarp plugin in Fiji. For better visualization, the pseudocolor look-up-table (LUT) was changed in the [13C6]glucose and [15N]amido-glutamine co-infused samples.
MYC signature in PDXs
Based on the microarray expression data from the PDXs used26,67 we downloaded the c6 oncogenic signatures from the Molecular Signatures Database68,69 and applied the GSVA package, v.1.48.3 (ref. 70) to infer sample specific MYC pathway activation. As the PDX cohort might not represent all types of breast cancers, we included the 1,980 METABRIC samples from elswehere37 and quantile-normalized them together before converting the expression values into z scores. For pathways that had a subset of genes that should be upregulated and another subset that should be downregulated, the final score was obtained as upregulated − downregulated. We compared the scores with the log-intensity expression with a scatter-plot.
Metabolic pathway analysis
Global feature extraction was carried out using Progenesis QI (Nonlinear Dynamics) tool following parameters used for the LC–MS analysis method (FWHM, 70,000, minimum chromatographic peak width of 0.166 min, min. intensity threshold of 105). Features with coefficient of variation below <30% across replicates from at least one class of the WMhigh, WMlow or WMmix tumor samples were further used for untargeted metabolic pathway analysis using MetaboAnalyst v.5.0 (ref. 71). A feature table consisting of 1,562 input m/z features from polar and apolar sample analysis of both polarities were processed using the mummichog algorithm. A feature significance cutoff of 0.05 P value for a Student’s t-test between WMhigh and WMlow was used and enriched pathways were identified using Homo sapiens (KEGG) pathway library (mass accuracy threshold of 10 ppm). For calculation of the enrichment score for the identified pathway, the number of significant feature hits from individual pathways with fold change difference higher in either WMhigh or WMlow samples was estimated. The enrichment score is then calculated with a modified mummichog algorithm by the significant hits coming from individual sample types rather than all significant hits and by using Fisher’s exact test P value for pathway enrichment to scale the enrichment score. Enriched pathways that consist of pathway size >1 in the library were retained for the analysis.
Conditional probability for ion coexpression with WMhigh clones
The list of metabolites associated with enriched pathways in WMhigh tumors were compared to the DEFFI-MS data for deprotonated ions. Conditional probabilities P (metabolite level | myc level) were estimated from the multi-run merged normalized and batch-effect-corrected DEFFI data. Metabolite intensities were discretized into low, mid and high levels by thresholding to the t1 = 33th and t2 = 66th percentiles (y ≤ t1⇒low, t1 < y ≤ t2⇒mid, y > t2⇒high). The conditional probabilities were calculated as a fraction of pixels with either low, mid or high intensity, stratified by MYC level. Only pixels from WMlow and WMhigh were used for the estimation. The calculated conditional probabilities were represented as a heat map using R v.3.6.2 (ref. 72) and the ComplexHeatmap v.2.2.0 (ref. 73) package.
METABRIC dataset analysis
Transcript levels of SLC5A6 were evaluated in human breast cancer samples from the publicly available METABRIC microarray gene expression dataset37. Samples were subsequently annotated with molecular subtype based on Prediction Analysis of Microarray 50 allocations74, estrogen receptor status, grade, size, number of lymph nodes positive and proliferation (expression of Aurora Kinase A). Thereafter, samples were ordered according to MYC signature expression (average transcript levels of 335 genes), retrieved from a previously published core MYC expression signature (consisting of 398 genes)75. Data analyses were undertaken using R v.3.6.1 (ref. 72), the ComplexHeatmap v.2.2.0 (ref. 73) and Circlize v.0.4.15 (ref. 76) packages.
Chip-seq
Publicly available data from Sabo et al.36 were downloaded from the Gene Expression Omnibus repository under accession no. GSE51011. An E-box rich region inside the murine Slc5a6 promoter was identified using the genomic sequence extracted from the Ensembl database and the respective region was identified in the Chip-seq raw data. The respective binding intensities of MYC to this region were plotted for pre-tumor as well as tumor cells.
Statistics
Pairwise comparisons were generally carried out using the Student’s t-test in Excel or using the R package ggplot2 v.3.4.0 and ggpubr v.0.4.0. No statistical methods were used to predetermine sample sizes but our sample sizes are similar to those reported in previous publications15,18.
When possible, mice from the same litter were randomized into groups. To avoid litter effects, different cell types (for example WMhigh, WMlow and WMmix) were implanted into mice from different litters in a random fashion. This was also the case in all studies with diet alteration. Before changing the diet, litters were mixed and randomized. Samples were randomized for metabolomics analysis. In preclinical model experiments, tumor size and mouse weight measurements were blinded as well as treatment regimes. DESI/DEFFI imaging was carried out blind and agnostic to the underlying tumor genotypes. Data analyses were not performed blind to the conditions of the experiments. Data were only excluded due to technical failure (for example over confluence, at start of an experiment, bad melting curve in qRT–PCR). No data were systematically excluded. Only mice with failed tumor grafting or tumor-unrelated health issues were excluded. Mice with failed metabolite administration due to improper cannulation and mice with failed tumor grafting were excluded.
For the NanoSIMS study with calcium pantothenate (dual label 13C and 15N), the 13C trace was recorded, but not analyzed, as it failed to show a signal above background, due to a higher natural abundance of 13C compared to 15N.
Statistical methods embedded in R-code-based image analysis, such as the ion colocalization analysis and the correlation dendrogram, are indicated in the respective sections.
All box-and-whisker plots represent the following: line, median; box, IQR; whiskers, 1.5 × IQR limited by largest/smallest NEV.
Data distribution was assumed to be normal but this was not formally tested.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.